Strontium »
PDB 1cs7-2grb »
1vc0 »
Strontium in PDB 1vc0: Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution
Protein crystallography data
The structure of Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution, PDB code: 1vc0
was solved by
A.Ke,
K.Zhou,
F.Ding,
J.H.D.Cate,
J.A.Doudna,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.53 / 2.50 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 108.944, 108.944, 191.754, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 24.6 / 27.3 |
Strontium Binding Sites:
The binding sites of Strontium atom in the Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution
(pdb code 1vc0). This binding sites where shown within
5.0 Angstroms radius around Strontium atom.
In total 2 binding sites of Strontium where determined in the Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution, PDB code: 1vc0:
Jump to Strontium binding site number: 1; 2;
In total 2 binding sites of Strontium where determined in the Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution, PDB code: 1vc0:
Jump to Strontium binding site number: 1; 2;
Strontium binding site 1 out of 2 in 1vc0
Go back to
Strontium binding site 1 out
of 2 in the Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution
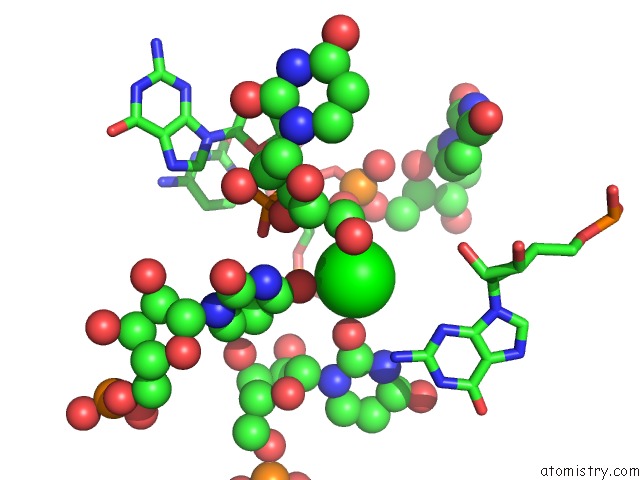
Mono view
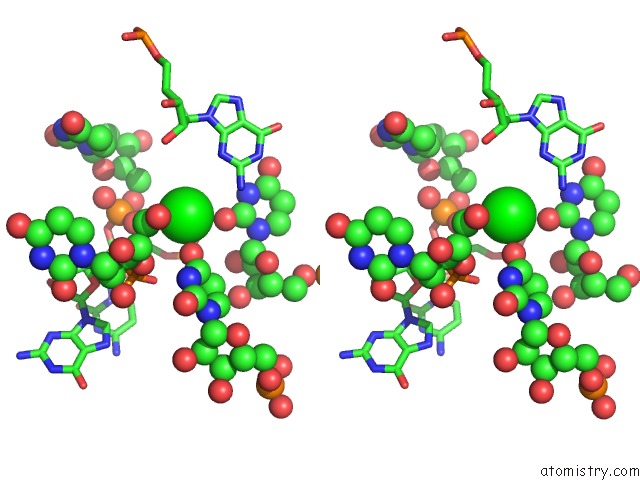
Stereo pair view

Mono view

Stereo pair view
A full contact list of Strontium with other atoms in the Sr binding
site number 1 of Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution within 5.0Å range:
|
Strontium binding site 2 out of 2 in 1vc0
Go back to
Strontium binding site 2 out
of 2 in the Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution
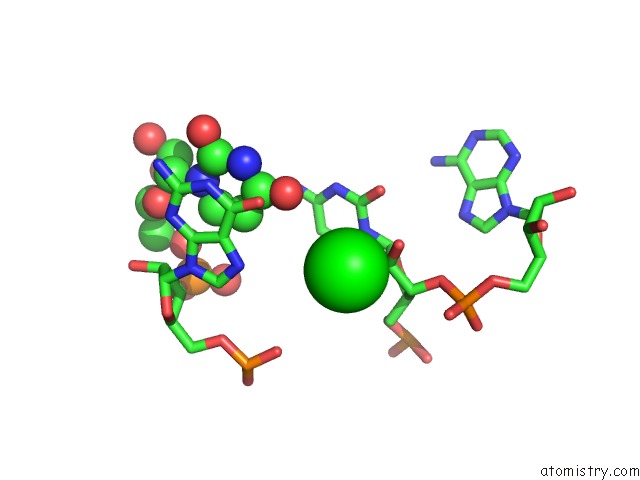
Mono view
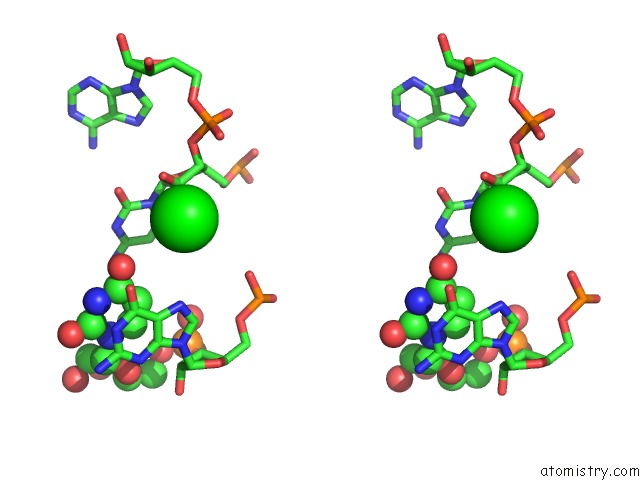
Stereo pair view

Mono view

Stereo pair view
A full contact list of Strontium with other atoms in the Sr binding
site number 2 of Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U Mutaion, in Imidazole and SR2+ Solution within 5.0Å range:
|
Reference:
A.Ke,
K.Zhou,
F.Ding,
J.H.D.Cate,
J.A.Doudna.
A Conformational Switch Controls Hepatitis Delta Virus Ribozyme Catalysis Nature V. 429 201 2004.
ISSN: ISSN 0028-0836
PubMed: 15141216
DOI: 10.1038/NATURE02522
Page generated: Thu Oct 10 14:06:39 2024
ISSN: ISSN 0028-0836
PubMed: 15141216
DOI: 10.1038/NATURE02522
Last articles
Cl in 8GFACl in 8GK4
Cl in 8GJT
Cl in 8GDK
Cl in 8GHH
Cl in 8GEQ
Cl in 8GEO
Cl in 8GDJ
Cl in 8GD5
Cl in 8GD3